| No. | Result | Comment |
|---|
| 1. |
Screening/Task Flowchart | 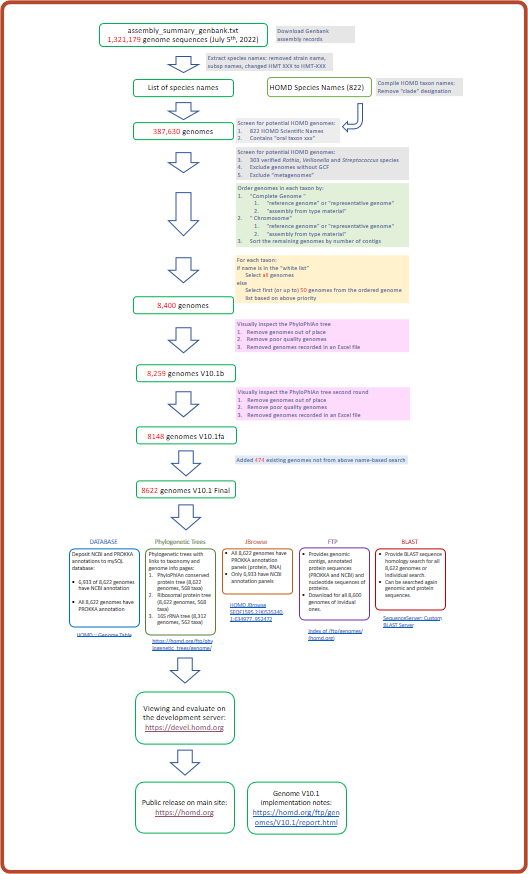 |
| 2. |
PhyloPhlan Tree of initial 8,400 genomes | Curation Notes in Excel |
| 3. | PhyloPhlan Tree of 8,259 genomes after first pass V10.1b | |
| | PhyloPhlan Tree of 8,259 genomes after 2nd pass V10.1c | added isolate info, numbered all genomes, so when scrolling we know how deep we are in the tree |
| | PhyloPhlan Tree of 8,173 genomes after 3rd pass V10.1d | dropped 86 genomes curated by Floyd, see red GCA IDs in Column E of this Excel file |
| | PhyloPhlan Tree of 8,146 genomes after 4th pass V10.1e | additional 27 misnamed genomes in the S. oralis and S. infantis clades were dropped see this Excel file |
| | PhyloPhlan Tree of 8,148 genomes V10.1fa | added GCA_000700805.1 and GCA_900459175.1 requested in the above Excel file |
| | PhyloPhlan Tree of 8,622 genomes V10.1fa | added 474 existing genomes (SEQFs) that did not come up from above name-based search |
| 4. |
Tasks performed for testing:
1. Deposit both NCBI and PROKKA annotations into HOME Genome Database (Andy)
2. Compile phylogenetic trees for:
a. Conversed protein tree (Phylophlan) (George)
b. 16S rRNA extracted from all genomes (George)
c. Ribosomal protein tree (George)
3. Compile BLAST data files for BLAST search (George)
4. Compile NCBI and PROKKA download files on FTP (George)
5. Render Jbrowse viewing for all genomes (George)
|
| 5. |
Testing and evaluation:
1. Deposit both NCBI and PROKKA annotations into HOME Genome Database (Andy) - test search speed and result
2. Compile phylogenetic trees for:
a. Conversed protein tree (Phylophlan) (George)
b. 16S rRNA extracted from all genomes (George)
c. Ribosomal protein tree (George)
=> test load speed and links
3. Compile BLAST data files for BLAST search (George & Andy) - test search performance (speed)
4. Compile NCBI and PROKKA download files on FTP (George) - test download
5. Render Jbrowse viewing for all genomes (George) - test links and browsing experience.
|
6. |
Publis release of V10.1
1. HOMD V3.1 = 16S RefSeq 15.22 + Genome V10.1
2. Point the public homd.org to devel.homd.org
3. Place this report in the update history
|